Single-Cell Analysis for Alzheimer’s: Integrating Transcriptomics and Synthetic Biology
Authors: Nanjundeshwara N., Lakshmiah A., Raghavan R.; Awareness 2025; 2(1); 8-20.
- Jan 06, 2025
- 115 views

Abstract
Alzheimer’s disease (AD) is a complex neurodegenerative disorder characterised by progressive cognitive decline. Single-cell transcriptomics has unveiled the complex cellular landscape of Alzheimer’s disease (AD), revealing distinct disease-associated cell states and gene expression profiles. This information provides a crucial foundation for leveraging the power of synthetic biology to develop targeted interventions. This paper explores the integration of these two fields, focusing on applications such as: 1) using transcriptomic data to design cell-type-specific synthetic constructs; 2) modelling and manipulating disease-relevant cell-cell interactions; and 3) engineering therapeutic cells for targeted drug delivery or immunomodulation. We discuss the potential of this integrated approach to advance our understanding of AD pathogenesis and pave the way for innovative therapeutic strategies, while also considering the associated challenges and future directions.

Graphical Abstract: Spatial Omics and Gene Circuits complement each other to overcome challenges and
limitations.
Full Text
1. Introduction
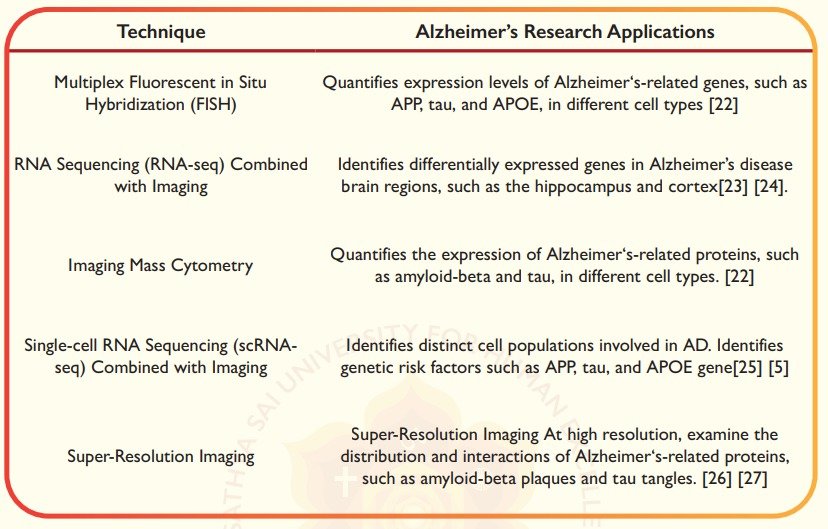
Impaired neurogenesis contributes to neurodegenerative diseases such as Alzheimer’s, Parkinson’s, Huntington’s, and Lewy Body dementia in several ways. Amyloid β protein (Aβ) in neuritic plaques of Alzheimer’s disease (AD) has been considered the molecular driver of Alzheimer’s pathogenesis and progression [1]. Spatial omics technologies, such as spatial transcriptomics (ST) and spatial isoform transcriptomics, provide a means of studying gene expression in tissue architecture and spatial organisation [2]. For example, ST studies have identified disease-associated microglia, which exhibit unique gene expression profiles and functional properties compared to homeostatic microglia. Similarly, distinct subtypes of astrocytes and neurons with altered gene expression patterns have been identified in AD brains.
Single-cell multi-omics technologies enable direct measurement of cell-specific responses, perturbations, and signalling pathways in disease pathogenesis. This provides insights into cellular spatial location, gene expression variations, genetic risk factors like APOE4, and single-cell or subcellular patterns with high spatial resolution. [3]. Spatial isoform transcriptomics (SiT) enables explicitly the characterisation of isoform expression and sequence heterogeneity at a spatial resolution using long-read sequencing. Spatial isoform transcriptomics involves characterising regional isoform switching and differential isoform usage for genes related to specific functions like brain activity [4]. Recent studies have identified rare subsets of immune cells infiltrating the brain in AD, which may contribute to neuroinflammation.
Advances in spatial transcriptomics, such as the 10x Genomics Visium platform, have provided detailed molecular maps that surpass the limitations of single-cell RNA sequencing methods [5]. ST platforms has enabled sub-regional gene expression analysis and comparable sequencing depth for gene number per spot [6]. Developments in super-resolution and expansion microscopy offer higher spatial resolution in omics, enabling visualization of cellular details and interactions crucial for understanding neural circuits.
Table 1. Examples of Spatial Omics Techniques in Alzheimer’s Research
2. Synthetic Biology for AD Research:
Synthetic biology engineer cells or organisms to recapitulate AD hallmarks like Aβ aggregation, tau hyperphosphorylation, and neuroinflammation. Gene circuit technology controls gene expression in response to signals, enabling the design of circuits that mimic or modulate AD pathways. Here are some synthetic biology applications:
2.1 Designer Receptors Exclusively Activated by Designer Drugs (DREADDs)
DREADDs are engineered G protein-coupled receptors (GPCRs) that are selectively activated by synthetic ligands (like CNO and clozapine-N-oxide). Chemogenetic manipulation of astrocyte activity using DREADDs can improve neuronal function and behaviour in normal animals and disease models. Different DREADD variants (e.g., hM3Dq for activation, hM4Di for inhibition) can activate or inhibit specific neuronal populations in specific cell types. They allow researchers to control neuronal activity with high temporal and cell-type specificity remotely [8] [9].
2.2 BRANEnet BRANEnet
(Brain Research through Advancing Neurotechnologies) is leveraging advanced molecular genetics techniques. The Alzheimer’s disease risk gene BIN1 is critical in regulating calcium homeostasis, electrical activity, and gene expression in glutamatergic neurons. BRANEnet utilises single-cell RNA sequencing, epigenomic analyses, and genome-wide association studies to provide deeper insights into AD [10]. Studies have shown that BIN1 knockout human-induced neurons (hiNs) exhibit reduced activity-dependent internalisation and higher expression of the L-type voltage-gated calcium channel Cav1.2, which affects neuronal calcium transients and electrical activity[11]. The insights gained from circuit omics can inform the design of gene therapies targeted to specific neural circuits, such as the entorhinal cortex-hippocampal (EC-HPC) system in AD patients [12]. BRANEnet aids in understanding microglia, immune cells, and the impact of genetic risk factors like APOE mutations. By integrating multi-omics data, BRANEnet outperforms baseline methods in tasks like transcription factor target prediction and network inference. Using a random walk-based Positive Pointwise Mutual Information matrix, BRANEnet captures relevant context for understanding complex neural interactions [13].
2.3 Viral Delivery Systems
Contemporary viral tracing strategies and activity recording methods investigate circuit architecture and function. The rapid advancement of CRISPR and anti-CRISPR tools enhances the robustness and applicability of synthetic biology systems. This system includes a genetic circuit that consists of libraries of guide RNAs, synthetic operators, transcriptional activators, and other regulatory elements [14]. Synthetic gene circuits enable permanent neural ensemble labelling, crucial for understanding memory dynamics. Designer cells, autonomous closed-loop therapeutic cells, avoid repeated inducer administration. Genetic devices like toggle switches and repressilators regulate cellular processes. Designer cells with sensing modules detect disease markers and trigger therapeutic agent secretion without external inducers [15] [16].
Optogenetics combines optics and genetics to control specific neurons using photosensitive proteins encoded by adenoviruses. This technique allows for precise ion flow control across cell membranes, leading to inhibition or activation effects [17]. Intersectional genetic approaches and activity-dependent recombinases target specific neuronal populations based on molecular markers or activity patterns, allowing for more refined manipulation of neural circuits [18]. Advances in spatial omics and neuroscience, including optogenetics and synthetic biology, continue to deepen our understanding and ability to manipulate these vital cells for therapeutic purposes [19] [20]. Pseudotyped rabies virus maps monosynaptic connections with cell-type specificity. Genetically encoded indicators reveal neural circuit function in awake animals. Trans-neuronal tracing techniques like HSV-1 H129 and rabies virus map cell-type-specific AD circuits. Multiplexed mapping and sequencing (MAPseq, BRICseq) refine connectivity analysis with single-cell resolution. [15].
2.4 E-SARE and RAM Synthetic Promoters
The RAM promoter enhances IEG labelling for better neuronal activity tracking. Synthetic promoters like E-SARE boost neuronal activation in vivo, aiding neural circuit studies. These promoters can create biosensors and complex circuits with novel functions, advancing our understanding of memory and behaviour [21].
3. Rationale for Integrating Transcriptomics and Synthetic Biology:
Transcriptomic data can inform the design of synthetic constructs that target specific cell types or pathways implicated in AD, leading to more precise and effective interventions. Additionally, an integrated systems-level understanding of AD-associated neural circuit mechanisms requires new multimodal and multi-scale interrogations to study and treat circuits vulnerable to AD. Transcriptomics can reveal how different cell types communicate in the AD brain. At the same time, synthetic biology can be used to engineer model systems to study these interactions in a controlled manner [28] [29].
3.1 Synthetic receptor systems
Synthetic receptors are potent tools in cell and gene therapies, allowing for precise control of therapeutic cells and genetic modules. These receptors can regulate the production of bioactive payloads by sensing and processing user-defined signals or biomarkers. Examples of synthetic receptor systems include chimeric antigen receptors (CARs) and synthetic Notch (synNotch) receptors [30] [31].
3.2 Advantages over traditional methods:
- Specificity: Synthetic receptors offer high specificity for their ligands, minimising off-target effects.
- Temporal control: The timing of receptor activation can be precisely controlled.
- Modularity: Synthetic receptors can be easily modified or combined to create new functionalities.
Synthetic receptors are used in targeted gene therapies and fundamental research to investigate cell signalling, differentiation, migration, and morphogenesis [32] [33]. They can program engineered cells to self-organise into multicellular structures or pattern three-dimensional tissues. Synthetic receptor systems like LOCa can modulate aberrant self-renewal of hematopoietic stem cells and mitigate neurodegeneration in models of Alzheimer’s disease [34]. Synthetic receptors can be partially or fully modular. Partially modular receptors retain the original sensor or actuator domain, while fully modular receptors have both engineered. Cell-surface receptors include enzyme-linked, G-protein-coupled, and ion channel-like receptors. [35].
Combining synthetic receptors and omics technologies offers unprecedented insights into cellular signalling, function, and disease. Techniques like TRAP enable targeted neuronal study based on activity, offering a dynamic approach to neural circuit research. [36]. Viral Tracers for Connectivity Mapping, both retrograde and anterograde, have been employed to map neural connections [16]. Integrating synthetic biology with single-cell analysis offers a comprehensive view of AD and enables targeted therapies. The E-SARE promoter shows higher reporter expression and dynamic range than natural IEG promoters. Combining synthetic promoters with omics technologies provides deeper insights into gene regulation and enables engineering new biological functions. [21].
Therapeutic strategies targeting astrocytes using Designer Receptors Exclusively Activated by Designer Drugs are being explored [35]. RNA sequencing can reveal changes in gene expression in response to DREADDmediated neuronal activation or inhibition. The effects of astrocyte manipulation vary depending on the specific DREADD receptor used. Other factors include targeted brain region, timing of intervention, and the heterogeneity of astrocytes in different brain regions and disease conditions [37]. DREADDs control neuronal activity but don’t directly reveal downstream molecular consequences. ATAC-seq reveals chromatin structure changes, indicating altered transcriptional regulation. Single-cell analysis informs cell-based therapies by identifying specific cell types for therapeutic agent delivery or immune modulation. [36].
3.3 Comprehensive Readout of Activity and Cell Type Markers (CRACK)
Combining transcriptomics, circuit tracing, and modulation offers a potent synthesis for studying diseasespecific circuitry. Optogenetics and chemogenetics enable precise neural activity modulation and subsequent molecular characterization. Optogenetics uses light-sensitive proteins to control neural activity, providing precise manipulation of specific cell types or neural circuits. [38]. On the other hand, CRACK, chemogenetics, offers a less invasive method than optogenetics for altering neural activity in genetically defined neurons of animals [39]. CRACK technology combines population calcium imaging with multiplexed in situ hybridization. It creates brain-wide projections of identified cells and their behavioural tuning properties, enabling the study of specific circuit functions in AD. [11].
4. Advantages of Integrating Spatial Omics with Gene Circuit Technology
Spatial omics technologies offer unbiased spatial profiling of the transcriptome, revealing the operation of gene switches within the brain’s complex tissue environment.
4.1 Context-Dependent Behaviour
Gene circuits modulate cellular function and adapt to regulatory networks. Circuit function varies with cellular context. Spatial omics reveal how circuit performance changes across cellular locations, enabling contextspecific optimization. Amyloid plaques and tau tangles, AD hallmarks, vary across brain regions. Spatial omics unravel tissue heterogeneity and the spatial context of molecular components, aiding in optimizing synthetic biological systems for various applications. [2].
4.2 Off-Target Effects on Cellular Interactions
Spatial data enables effective gene switch design. Lysate-based cell-free systems (CFS) are valuable synthetic biology tools but lack living cell properties. Spatial omics visualizes circuit impact on gene expression, informing the development of more sophisticated gene circuit designs by identifying off-target effects.[40].
4.3 Predictability
In Alzheimer’s, gene expression patterns likely change over time and across different brain regions as the disease progresses. Spatial transcriptomics coupled with time-series studies can help capture the spatiotemporal dynamics of gene regulation. However, ensuring the predictability of gene circuits remains a challenge, particularly for complex circuit functions [41]. Using orthogonal guide sequences, insulating DNA sequences, and whole-cell omics measurements can improve the predictability of gene circuits [42].
4.4 Resource Competition
Resource competition couplings pose a significant barrier to the successful engineering of cells across various organisms, including bacteria, fungi, and mammals [43]. This necessitates precise control and systematic modification of critical variables like recording environment shape and layout. Gene expression burden, diverting resources from natural functions, hinders cell growth and productivity. [44]. Spatial omics identifies areas of resource depletion due to circuit activity, optimizing circuit design. Identifying specific cell types and circuitry within the affected entorhinal-hippocampal system is crucial for developing targeted circuit therapies. [45]. Information theory assesses neural circuit pattern separation performance. Spatial omics reveals circuit effects on neighbouring genes, enabling disruption minimization. Gene-circuit approaches quantify stimulus or environmental shift scale changes through behavioural experiments [46].
4.5 Dynamic Process Control
Spatial transcriptomics will help study spatial and temporal selectivity governing diverse functions within the neocortex. For cell type-specific visualisation, the formation of learning-specific ephemeral and memoryspecific enduring synapses utilising gene engineering techniques was demonstrated [47].
5. Future Directions
Multi-omics algorithms integrate genomic, proteomic, metabolomic, and other biological data to offer tailored AD management and functional recovery solutions. Fluorescence imaging of endogenous proteins, like PSD-95 in neurons using super-resolution techniques, is being developed. Combining molecular dynamics simulations with machine learning models offers comprehensive insights. Ion channels, complex proteins undergoing conformational changes upon activation, are challenging to model accurately. Cryo-EM provides detailed structural insights, improving computational model accuracy and drug design efforts. [48]. Predictive algorithms, powered by multi-omics data analysis, are revolutionising the field of rehabilitation and assistive systems. Bio Nexus Sentinel software platform integrates cytohistological RNA-seq and bioregulatory network exploration [49]. The patient-in-the-loop framework represents a unifying approach that can bridge the gap between computational modelling and clinical practice in neurorehabilitation. We can significantly enhance assistive augmentative rehabilitation by prioritising multidisciplinary collaboration and leveraging cutting edge research [50].
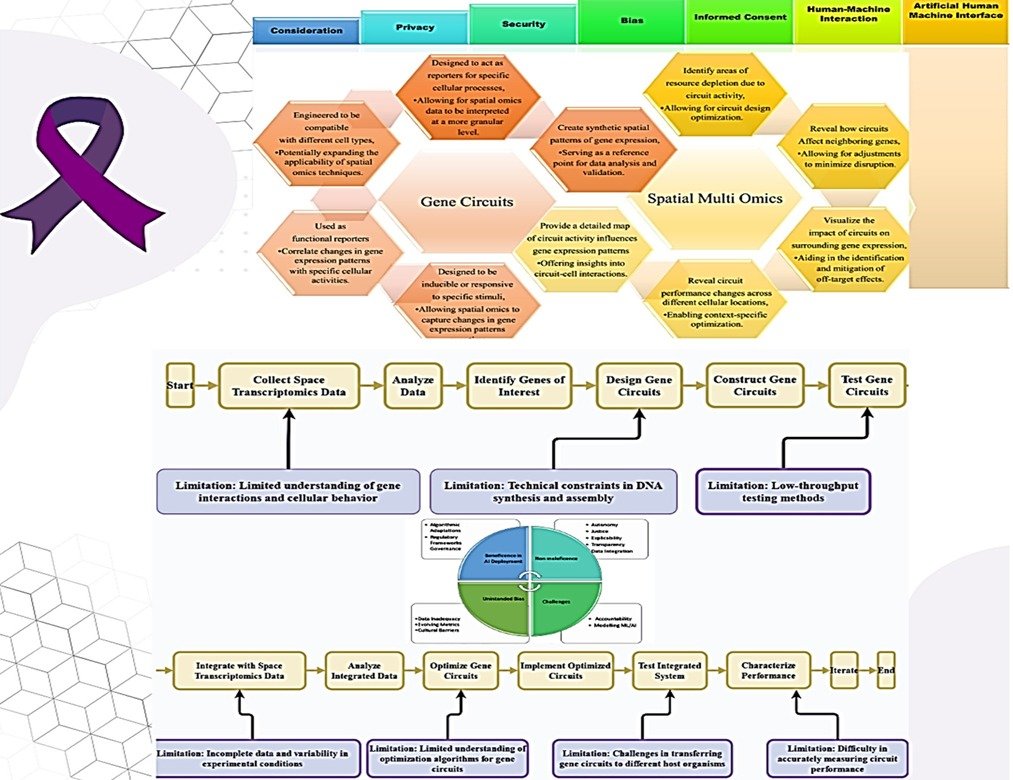
Figure 1. Synthetic Biology Solutions for Alzheimer’s Disease: Technological Limitations and Ethical Challenges [created with Microsoft PowerPoint and Canva]
5.1 Microfluidic Technology
Organ-on-a-chip (OOC) technology is a cutting-edge innovation that simulates the functions and physiology of human organs on a microfluidic chip [51][52]. Microfluidic organ chips and OOCs create a controlled microenvironment that mimics factors like fluid flow, mechanical forces, and chemical gradients, which are crucial for maintaining cell function and behaviour [53]. Integrating single-cell transcriptomics and synthetic biology can advance in vitro systems using microfluidic chips to enhance understanding and drug development. Additionally, microfluidic platforms integrate various omics data, improving understanding of cellular heterogeneity and interactions within complex biological systems [54].
5.2 Organoids
Patient-derived organoids enable personalized treatment. Biomimetic scaffolds promote tissue healing and reduce inflammation. Advanced drug-delivery materials improve treatment. Brain organoids are used for toxicology, disease modelling, infection studies, personalized medicine, and gene-environment interaction studies. Organoids advance AD drug screening. Patient-derived organoid models investigate pathogenesis, guide clinical treatment, and facilitate drug screening. Stem cell-derived and biomimetic scaffold-based organoids advance tissue engineering. Biomaterials create scaffolds for tissue regeneration. Multiomics and bioprinting evaluate pain medication efficacy and safety. Advanced imaging provides rich organoid information. Combining organoids and AI bridges experimental models and real-world clinical scenarios [55]. Omics and synthetic biology accelerate EV-based therapies. Digital microfluidics streamlines EV isolation for liquid biopsies.
5.3 Nanotechnology-based therapies
Nanotechnology strategies are discussed for enhancing drug delivery in brain disorders, including gene therapy, enzyme replacement therapy, and nano-assisted therapies like nano-immunotherapy and nano-gene therapy [56]. Successful drug discovery often requires the integration of various metal-based carriers and nanoparticles to reduce inflammation and pass through biological barriers and minimal systemic toxicity [57]. Nanotechnology, nanocarriers, nano immunotherapy and nano gold therapy improve drug delivery to specific sites, reducing systemic side effects [58].
5.4 Artificial Intelligence
The concept of Organoid Intelligence (OI) combines organoids with AI to model cognition and enable biological computing applications [59]. Computational techniques, such as virtual screening and molecular mechanics/ dynamics simulations, enhance CAR-NK cell-based immunotherapy. Machine learning-based approaches target minimising collateral damage to healthy tissues, and optimise treatment protocols [60]. DeepD3 Framework is an open deep learning-based framework for quantifying dendritic spines in microscopy data, neural networks trained on data from different sources and experimental conditions [61]. Neuromorphic computing approaches can help identify patterns and make predictions beyond traditional methods’ capability. By continuing to push the boundaries and fostering multidisciplinary integration, researchers can make significant strides towards conquering Alzheimer’s disease.
6. Ethical Considerations
Recent advancements in transcriptomics and synthetic biology offer promising avenues for understanding and addressing AD. The multiomics and synthetic biology tools needs to address ethical considerations related to potential therapeutic applications. Careful monitoring and follow-up of patients in clinical trials are essential. Efforts should be made to ensure that these therapies are available to all patients who need them, regardless of their socioeconomic status.
- Off-target effects: Synthetic constructs may have unintended effects on other cellular processes or tissues. Thorough preclinical testing is crucial to minimise these risks.
- Immunogenicity: Engineered cells or gene therapies may trigger unwanted immune responses. Strategies to minimise immunogenicity has be developed [51][52]
- Informed Consent and Patient Autonomy: Synthetic biology is a complex field, and it may be difficult for patients to understand the risks and benefits of these therapies fully. Clear and accessible information should be provided to patients to ensure informed consent.
AD affects cognitive function, there are concerns about a patient’s ability to provide informed consent, especially as the disease progresses. Several ethical principles are laid out to avoid any unintended outcomes of synthetic biology applications (Figure 1).
7. Conclusion
Alzheimer’s disease is a complex neurodegenerative disorder characterised by progressive cognitive decline and memory loss. Transcriptomics, the study of gene expression, provides insights into the molecular mechanisms underlying AD. At the same time, synthetic biology enables the engineering of biological systems for therapeutic applications. Integrating spatial omics with synthetic gene circuit switches allows gene regulation within tissues in spatial and temporal contexts. Integrating allows overcoming individual technological limitations to showcase tissue atlas, with the intricate interplay of genes across different cellular subregions. Gene circuits illuminate the function, and spatial omics pinpoint the differentially expressed genes underlying the observed patterns [62]
Genetic circuits and spatial omics complement each other to provide a detailed spatial-temporal map of gene expression patterns, offering novel insights into neuroscience. Combining these two fields allows us to understand AD pathogenesis better, identify novel therapeutic targets, and develop innovative treatment strategies.
Conflicts of Interest: There are no conflicts of interest to disclose.
Data Availability Statement: No new data were created during this study.
Data sharing: does not apply to this article.
Funding: This research received no external funding.
Institutional Review Board Statement: Not applicable.
Informed Consent Statement: Not applicable
References
- Zhang Y, Chen H, Li R, Sterling K, Song W (2023) Amyloid β-based therapy for Alzheimer’s disease: challenges, successes and future. Sig Transduct Target Ther 8:1–26. https://doi.org/10.1038/s41392-023- 01484-7
- Park J, Kim J, Lewy T, Rice CM, Elemento O, Rendeiro AF, Mason CE (2022) Spatial omics technologies at multimodal and single cell/subcellular level. Genome Biol 23:1–19. https://doi.org/10.1186/s13059-022- 02824-6
- Zhang X, Lü P, Shen X (2022) Applications of single-cell multi-omics sequencing in deep understanding of brain diseases. Clinical and translational discovery. https://doi.org/10.1002/ctd2.95
- Lebrigand K, Bergenstråhle J, Thrane K, Mollbrink A, Meletis K, Barbry P, Waldmann R, Lundeberg J (2023) The spatial landscape of gene expression isoforms in tissue sections. Nucleic Acids Res 51:e47. https://doi. org/10.1093/nar/gkad169
- (2024) Spatial Gene Expression. In: 10x Genomics. https://www.10xgenomics.com/products/spatial-geneexpression
- Vanrobaeys Y, Peterson ZJ, Walsh EN, Chatterjee S, Lin L-C, Lyons LC, Nickl-Jockschat T, Abel T (2023) Spatial transcriptomics reveals unique gene expression changes in different brain regions after sleep deprivation. Nat Commun 14:7095. https://doi.org/10.1038/s41467-023-42751-z
- Hofbauer B, Zandawala M, Reinhard N, Rieger D, Werner C, Evers JF, Wegener C (2024) The neuropeptide pigment-dispersing factor signals independently of Bruchpilot-labelled active zones in daily remodelled terminals of Drosophila clock neurons. European Journal of Neuroscience. https://doi.org/10.1111/ ejn.16294
- Lim J, Souiki A, Ahmad P, Oomen CA, Huis in ’t Veld G, Lansink CS, Pennartz CMA, Olcese U (2024) Transient DREADD manipulation of the dorsal Dentate Gyrus in rats impairs disambiguation of similar place-outcome associations. https://doi.org/10.1101/2024.02.20.581173
- Albrecht A, Müller I, Weiglein A, Pollali E, Çalışkan G, Stork O (2022) Choosing memory retrieval strategies: A critical role for inhibition in the dentate gyrus. Neurobiology of Stress 20:100474. https://doi. org/10.1016/j.ynstr.2022.100474
- Kiessling P, Kuppe C (2024) Spatial multi-omics: novel tools to study the complexity of cardiovascular diseases. Genome Med 16:1–17. https://doi.org/10.1186/s13073-024-01282-y
- Grieco SF, Holmes TC, Xu X (2023) Probing neural circuit mechanisms in Alzheimer’s disease using novel technologies. Mol Psychiatry 28:4407–4420. https://doi.org/10.1038/s41380-023-02018-x
- Igarashi KM (2022) Entorhinal cortex dysfunction in Alzheimer’s disease. Trends in Neurosciences 46:124– 136. https://doi.org/10.1016/j.tins.2022.11.006
- Jagtap S, Pirayre A, Bidard F, Duval L, Malliaros FD (2022) BRANEnet: embedding multilayer networks for omics data integration. BMC Bioinformatics 23:429. https://doi.org/10.1186/s12859-022-04955-w
- Du P, Lou C, Zhao X, Wang Q, Ji X, Wei W (2021) CRISPR-Based Genetic Switches and Other Complex Circuits: Research and Application. Life 11:1255. https://doi.org/10.3390/life11111255
- Bouin A, Wu G, Koyuncu OO, Ye Q, Kim K-Y, Wu MY, Tong L, Chen L, Phan S, Mackey MR, Ramachandra R, Ellisman MH, Holmes TC, Semler BL, Xu X (2024) New rabies viral resources for multi-scale neural circuit mapping. Molecular Psychiatry 1–17. https://doi.org/10.1038/s41380-024-02451-6
- Du W, Li E, Guo J, Arano R, Kim Y, Chen Y-T, Thompson A, Oh SJ, Samuel A, Li Y, Oyibo HK, Xu W (2023) Directed stepwise tracing of polysynaptic neuronal circuits with replication-deficient pseudorabies virus. Cell Rep Methods 3:100506. https://doi.org/10.1016/j.crmeth.2023.100506
- Zhang Q, Li T, Xu M, Islam B, Wang J (2024) Application of Optogenetics in Neurodegenerative Diseases. Cell Mol Neurobiol 44:57. https://doi.org/10.1007/s10571-024-01486-1
- Swanson JL, Chin P-S, Romero JM, Srivastava S, Ortiz-Guzman J, Hunt PJ, Arenkiel BR (2022) Advancements in the Quest to Map, Monitor, and Manipulate Neural Circuitry. Frontiers in Neural Circuits 16:886302. https://doi.org/10.3389/fncir.2022.886302
- Zhu D, Johnson HJ, Chen J, Schaffer DV (2022) Optogenetic Application to Investigating Cell Behavior and Neurological Disease. Frontiers in Cellular Neuroscience 16:811493. https://doi.org/10.3389/ fncel.2022.811493
- Benisch M, Aoki SK, Khammash M (2023) Unlocking the potential of optogenetics in microbial applications. Current Opinion in Microbiology 77:102404. https://doi.org/10.1016/j.mib.2023.102404
- Pang B, Wu X, Chen H, Yan Y, Du Z, Yu Z, Yang X, Wang W, Lu K (2024) Exploring the memory: existing activity-dependent tools to tag and manipulate engram cells. Frontiers in Cellular Neuroscience 17:1279032. https://doi.org/10.3389/fncel.2023.1279032
- Xu P, Peng J, Yuan T, Chen Z, He H, Wu Z, Li T, Li X, Wang L, Gao L, Yan J, Wei W, Li CT, Luo Z-G, Chen Y (2024) High-throughput mapping of single-neuron projection and molecular features by retrograde barcoded labeling. eLife 13:e85419. https://doi.org/10.7554/elife.85419
- . Yang F, Zhao Z, Zhang D, Xiong Y, Dong X, Wang Y, Yang M, Pan T, Liu C, Liu K, Lin Y, Liu Y, Tu Q, Dang Y, Xia M, Mi D, Zhou W, Xu Z (2024) Single-cell multi-omics analysis of lineage development and spatial organization in the human fetal cerebellum. Cell Discovery 10:22. https://doi.org/10.1038/s41421-024- 00656-1
- Gao M-Y, Wang J-Q, He J, Gao R, Zhang Y, Li X (2023) Single-Cell RNA-Sequencing in Astrocyte Development, Heterogeneity, and Disease. Cellular and Molecular Neurobiology 43:3449–3464. https:// doi.org/10.1007/s10571-023-01397-7
- Shi Y, Huang L, Dong H, Yang M, Ding W, Zhou X, Lu T, Liu Z, Zhou X, Wang M, Zeng B, Sun Y, Zhong S, Wang B, Wang W, Yin C, Wang X, Wu Q (2024) Decoding the spatiotemporal regulation of transcription factors during human spinal cord development. Cell Research 34:193–213. https://doi.org/10.1038/s41422- 023-00897-x
- (2024) nCounter® Analysis Systems for Biomarker Validation and Biomarker Development. In: NanoString. https://nanostring.com/products/ncounter-analysis-system/ncounter-systems-overview/
- Cs C, J X, Y S, Sr P, Je H, M K, G J, Hm K, Jh L (2021) Microscopic examination of spatial transcriptome using Seq-Scope. Cell 184:. https://doi.org/10.1016/j.cell.2021.05.010
- Siddiqui M, Tous C, Wong WW (2022) Small molecule-inducible gene regulatory systems in mammalian cells: progress and design principles. Current Opinion in Biotechnology 78:102823. https://doi.org/10.1016/j. copbio.2022.102823
- Eisenhut P, Marx N, Borsi G, Papež M, Ruggeri C, Baumann M, Borth N (2024) Manipulating gene expression levels in mammalian cell factories: An outline of synthetic molecular toolboxes to achieve multiplexed control. New Biotechnology 79:1–19. https://doi.org/10.1016/j.nbt.2023.11.003
- Barbier I, Kusumawardhani H, Chauhan L, Harlapur PV, Jolly MK, Schaerli Y (2023) Synthetic Gene Circuits Combining CRISPR Interference and CRISPR Activation in E. coli: Importance of Equal Guide RNA Binding Affinities to Avoid Context-Dependent Effects. ACS Synth Biol 12:3064–3071. https://doi.org/10.1021/ acssynbio.3c00375
- Kraus C, Sontheimer EJ (2023) Applications of Anti-CRISPR Proteins in Genome Editing and Biotechnology. Journal of Molecular Biology 435:168120. https://doi.org/10.1016/j.jmb.2023.168120
- Chen WCW, Gaidukov L, Lai Y, Wu M-R, Cao J, Gutbrod MJ, Choi GCG, Utomo RP, Chen Y-C, Wroblewska L, Kellis M, Zhang L, Weiss R, Lu TK (2022) A synthetic transcription platform for programmable gene expression in mammalian cells. Nat Commun 13:6167. https://doi.org/10.1038/s41467-022-33287-9
- Rajasekaran R, Chang C-C, Weix EWZ, Galateo TM, Coyle SM (2024) A programmable reaction-diffusion system for spatiotemporal cell signaling circuit design. Cell 187:345-359.e16. https://doi.org/10.1016/j. cell.2023.12.007
- Teng F, Cui T, Zhou L, Gao Q, Zhou Q, Li W (2024) Programmable synthetic receptors: the next-generation of cell and gene therapies. Signal Transduction and Targeted Therapy 9:7. https://doi.org/10.1038/s41392- 023-01680-5
- Lee H-G, Wheeler MA, Quintana FJ (2022) Function and therapeutic value of astrocytes in neurological diseases. Nat Rev Drug Discov 21:339–358. https://doi.org/10.1038/s41573-022-00390-x
- Kim D, Tran A, Kim HJ, Lin Y, Yang JYH, Yang P (2023) Gene regulatory network reconstruction: harnessing the power of single-cell multi-omic data. npj Syst Biol Appl 9:1–13. https://doi.org/10.1038/s41540-023- 00312-6
- Pereira MJ, Ayana R, Holt MG, Arckens L (2023) Chemogenetic manipulation of astrocyte activity at the synapse— a gateway to manage brain disease. Front Cell Dev Biol 11:. https://doi.org/10.3389/ fcell.2023.1193130
- Ördög B, De Coster T, Dekker SO, Bart CI, Zhang J, Boink GJJ, Bax WH, Deng S, den Ouden BL, de Vries AAF, Pijnappels DA (2023) Opto-electronic feedback control of membrane potential for real-time control of action potentials. Cell Rep Methods 3:100671. https://doi.org/10.1016/j.crmeth.2023.100671
- Tan P, He L, Huang Y, Zhou Y (2022) Optophysiology: Illuminating cell physiology with optogenetics. Physiol Rev 102:1263–1325. https://doi.org/10.1152/physrev.00021.2021
- Wagner L, Jules M, Borkowski O (2023) What remains from living cells in bacterial lysate-based cell-free systems. Computational and Structural Biotechnology Journal 21:3173–3182. https://doi.org/10.1016/j. csbj.2023.05.025
- Şimşek E, Yao Y, Lee D, You L (2022) Toward predictive engineering of gene circuits. Trends in Biotechnology 41:760–768. https://doi.org/10.1016/j.tibtech.2022.11.001
- Dwijayanti A, Zhang C, Poh CL, Lautier T (2022) Toward Multiplexed Optogenetic Circuits. Frontiers in Bioengineering and Biotechnology 9:
- McBride CD, Grunberg TW, Del Vecchio D (2021) Design of genetic circuits that are robust to resource competition. Current Opinion in Systems Biology 28:100357. https://doi.org/10.1016/j.coisb.2021.100357
- Sequeiros C, Vázquez C, Banga JR, Otero-Muras I (2023) Automated Design of Synthetic Gene Circuits in the Presence of Molecular Noise. ACS Synthetic Biology 12:2865–2876. https://doi.org/10.1021/ acssynbio.3c00033
- Borzello M, Ramirez S, Treves A, Lee I, Scharfman H, Stark C, Knierim JJ, Rangel LM (2023) Assessments of dentate gyrus function: discoveries and debates. Nat Rev Neurosci 24:502–517. https://doi.org/10.1038/ s41583-023-00710-z
- Wang M, Song W-M, Ming C, Wang Q, Zhou X, Xu P, Krek A, Yoon Y-J, Ho L, Orr ME, Yuan G-C, Zhang B (2022) Guidelines for bioinformatics of single-cell sequencing data analysis in Alzheimer’s disease: review, recommendation, implementation and application. Molecular Neurodegeneration. https://doi.org/10.1186/ s13024-022-00517-z
- Severino L, Kim J, Nam M-H, McHugh TJ (2024) From synapses to circuits: What mouse models have taught us about how autism spectrum disorder impacts hippocampal function. Neuroscience & Biobehavioral Reviews 158:105559. https://doi.org/10.1016/j.neubiorev.2024.105559
- Torres R, Thal LB, McBride JR, Cohen BE, Rosenthal SJ (2024) Quantum Dot Fluorescent Imaging: Using Atomic Structure Correlation Studies to Improve Photophysical Properties. The Journal of Physical Chemistry C 128:3632–3640. https://doi.org/10.1021/acs.jpcc.3c07367
- Matzko RO, Konur S (2024) BioNexusSentinel: a visual tool for bioregulatory network and cytohistological RNA-Seq genetic expression profiling within the context of multicellular simulation research using ChatGPT-Augmented software engineering. Bioinformatics Advances vbae046. https://doi.org/10.1093/ bioadv/vbae046
- Cashaback JGA, Allen JL, Chou AH-Y, Lin DJ, Price MA, Secerovic NK, Song S, Zhang H, Miller HL (2024) NSF DARE—transforming modeling in neurorehabilitation: a patient-in-the-loop framework. Journal of NeuroEngineering and Rehabilitation 21:23. https://doi.org/10.1186/s12984-024-01318-9
- Dijk G, Poulkouras R, O’Connor RP (2022) Electroporation Microchip With Integrated Conducting Polymer Electrode Array for Highly Sensitive Impedance Measurement. IEEE Transactions on Biomedical Engineering 69:2363–2369. https://doi.org/10.1109/tbme.2022.3143542
- Habibey R, Arias JER, Striebel J, Busskamp V (2022) Microfluidics for Neuronal Cell and Circuit Engineering. Chemical Reviews 122:14842–14880. https://doi.org/10.1021/acs.chemrev.2c00212
- Sharkey C, White R, Finocchiaro M, Thomas J, Estevam J, Konry T (2024) Advancing Point-of-Care Applications with Droplet Microfluidics: From Single-Cell to Multicellular Analysis. Annual Review of Biomedical Engineering 26:. https://doi.org/10.1146/annurev-bioeng-110222-102142
- Yang N, Sun C, Dong C, Huang Y, Zhu Y, Gu Z (2024) Emerging microfluidics for the modeling and treatment of arthritis. Engineered Regeneration 5:153–169. https://doi.org/10.1016/j.engreg.2024.02.002
- Patel D, Shetty S, Acha C, Pantoja IEM, Zhao A, George D, Gracias DH (2024) Microinstrumentation for Brain Organoids. Advanced Healthcare Materials e2302456. https://doi.org/10.1002/adhm.202302456
- Dong H, Lin J, Tao Y, Jia Y, Sun L, Li WJ, Sun H (2024) AI-enhanced biomedical micro/nanorobots in microfluidics. Lab on a Chip 24:1419–1440. https://doi.org/10.1039/d3lc00909b
- Singh S, Sharma K, Sharma H (2024) Green Extracts with Metal-based Nanoparticles for Treating Inflammatory Diseases: A Review. Current Drug Delivery 21:544–570. https://doi.org/10.2174/156720182 0666230602164325
- 58. Kumar PPP, Mahajan R (2024) Gold Polymer Nanomaterials: A Promising Approach for Enhanced Biomolecular Imaging. Nanotheranostics 8:64–89. https://doi.org/10.7150/ntno.89087
- Hartung T, Pantoja IEM, Smirnova L (2024) Brain organoids and organoid intelligence from ethical, legal, and social points of view. Frontiers in Artificial Intelligence 6:1307613. https://doi.org/10.3389/ frai.2023.1307613
- Amaya-Rodriguez CA, Carvajal-Zamorano K, Bustos D, Alegría-Arcos M, Castillo K (2024) A journey from molecule to physiology and in silico tools for drug discovery targeting the transient receptor potential vanilloid type 1 (TRPV1) channel. Frontiers in Pharmacology 14:1251061. https://doi.org/10.3389/ fphar.2023.1251061
- Mahendrakar P, kumar D, Patil U (2024) A Comprehensive Review on MRI-based Knee Joint Segmentation and Analysis Techniques. Current Medical Imaging 20:e150523216894. https://doi.org/10.2174/157340562 0666230515090557
- Greiss F, Lardon N, Schütz L, Barak Y, Daube SS, Weinhold E, Noireaux V, Bar-Ziv R (2024) A genetic circuit on a single DNA molecule as an autonomous dissipative nanodevice. Nature communications 15:883. https://doi.org/10.1038/s41467-024-45186-2
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of SSSUHE and/or the editor(s). SSSUHE and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions, or products referred to in the content






